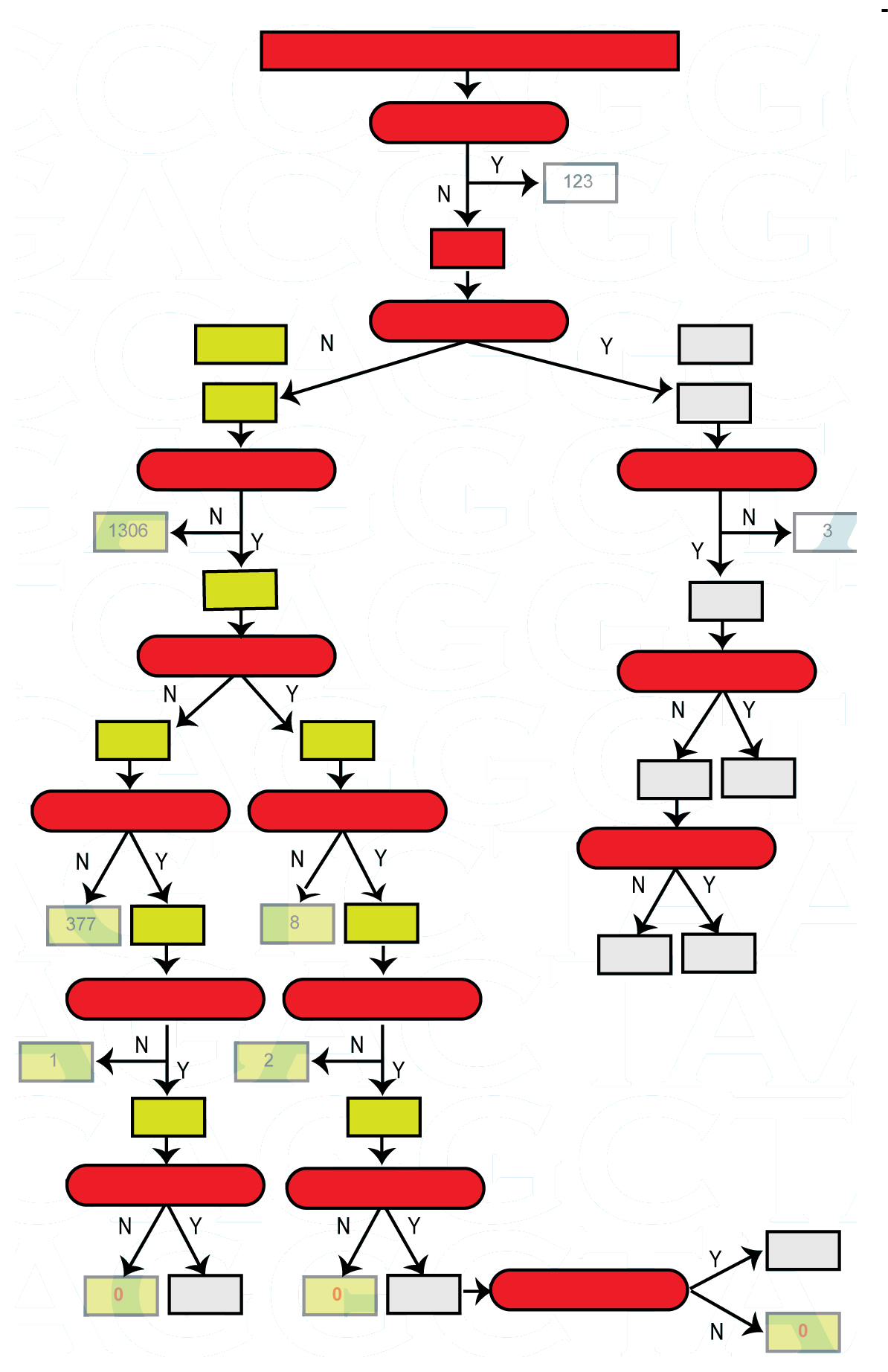
*** retired! only can download files. A new version is available here: FunSeq2
This site can be used to automatically score and annotate disease-causing potential of SNVs, particularly the non-coding ones. It can be used on cancer and personal genomes. It also contains a downloadable tool (found under 'Downloads').
Fun
ction based Prioritization ofSeq
uence Variants
Under 'Analysis', an online version of the tool is available, where a personal or cancer genome variant file (VCF or BED) can be uploaded and analysed.
Additionally, the tool can also detect recurrent annotation elements in non-coding regions when running with multiple genomes.